2. 郑州大学 合成生物学实验室,郑州 450001;
3. 北京化工大学 生命科学与技术学院,北京 100029
2. Laboratory of Synthetic Biology, Zhengzhou University, Zhengzhou 450001;
3. College of Life Science and Technology, Beijing University of Chemical Technology, Beijing 100029, China
中药黄芪(Astragali radix)是重要的临床大宗药材之一,为蒙古黄芪和膜荚黄芪的干燥根。研究表明,黄芪具有调节免疫、抗衰老、抗氧化、抗菌等多种药理活性[1]。黄芪含有复杂多样的天然产物,其中黄酮类、多糖类和皂苷类为主要的药用活性成分。黄芪中黄酮类活性成分主要包括黄酮、异黄酮、异黄烷、紫檀烷等,具有抗氧化[2]、抗肿瘤[3]、改善心血管功能[4]、预防骨质疏松[5]等药理作用。黄芪多糖主要有杂多糖、葡聚糖两大类,具有抗肿瘤[6]、抗炎症[7]、保护心脏[8]等药理活性。黄芪皂苷类成分包括Cycloartane型四环三萜、Oleanane型五环三萜及其他萜类[9]。黄芪甲苷的活性形式——环黄芪醇具有抑制脑缺血时脑细胞凋亡和神经炎症、维持血脑屏障[10]等药理作用。此外,环黄芪醇还是目前发现的唯一具有端粒酶活性的小分子萜类化合物,除了能够治疗神经退行性疾病[11],还具有抑制心肌纤维化[12]、增强抗肿瘤免疫[13]等作用,是抗衰老药物研发中的热门分子[14]。因此,黄芪已广泛应用于医药、日化、保健食品、禽畜饲料等多个领域。
环黄芪醇主要通过转化黄芪中的黄芪甲苷获取,但是现有的制备方法存在诸多不足,导致其难以低成本大规模生产。合成生物学方法可以帮助构建具有高产率、高纯度产物的可控生产平台,合成复杂多样的植物天然产物,并进一步揭示植物次生代谢合成途径及其关键酶[15]。目前,运用合成生物学方法已成功在酵母中异源合成了甘草次酸[16]、人参皂苷[17-19]、柴胡皂苷元[20]、罗汉果苷[21]等植物萜类天然产物,其中部分萜类天然产物的酵母合成已进行了产业化尝试,这为环黄芪醇在酵母等微生物中的合成提供了参考。本文综述了环黄芪醇的生物合成关键酶挖掘、酿酒酵母(Saccharomyces cerevisiae)底盘细胞改造,并对环黄芪醇的酵母高效合成进行了讨论。
1 环黄芪醇的现有制备方法环黄芪醇在黄芪中含量极低,难以直接获取。可以通过黄芪种植和组织培养技术获取黄芪甲苷,然后采用酸解法、Smith降解法、酶和微生物水解法等手段将黄芪甲苷转化为环黄芪醇。
酸解法可以水解黄芪甲苷生成环黄芪醇,常用硫酸、盐酸、乙酸在水或稀醇溶液中进行。楚治良等[22]将黄芪甲苷粉末加入到含有盐酸、甲醇和三氯甲烷的反应体系中,反应6天后,环黄芪醇的收率可达45%以上。Smith降解法是制备环黄芪醇的常用方法,使用高碘酸选择性氧化黄芪甲苷中的羟基,并进一步经硼氢化合物还原得到多糖醇,然后在酸性条件下多糖醇发生特异性水解,生成环黄芪醇。Feng等[23]采用Smith法降解黄芪甲苷制备环黄芪醇,对试剂用量优化后,环黄芪醇的转化产率可达80%以上。
酶和微生物水解法的原理是利用β-葡萄糖苷酶和β-木糖苷酶等多种糖基水解酶水解黄芪甲苷的多种糖苷键,从而得到环黄芪醇。Cheng等[24]筛选并克隆了Phycicoccus sp. Soil748(Bgps)中的β-葡萄糖苷酶,对该酶的最适温度和pH等酶学特性进行了研究,发现在45 ℃、pH7.0、底物质量浓度为80 mg/mL的条件下,该酶水解黄芪甲苷的转化率可达99.2%。Li等[25]从Dictyoglomus thermophilum中克隆纯化得到β-葡萄糖苷酶Dth3和β-木糖苷酶Xln-DT,并通过体外实验验证了酶活特性,在最优条件(75 ℃,pH5.5)下,Dth3和Xln-DT共催化可以在3 h内将1 g/L黄芪甲苷转化为0.63 g/L环黄芪醇,摩尔转化效率可达94.5%。Cheng等[26]在Pichia pastoris和Escherichia coli中进行密码子优化后表达了重组β-木糖苷酶Xyl-T,在特定的温度和pH下经过两步酶催化反应,该酶可以将20 g黄芪甲苷转化为12.02 g环黄芪醇,环黄芪醇收率达到96.5%。此外,双歧杆菌和乳酸菌等人体肠道菌和部分其他环境微生物也具有将黄芪甲苷转化为环黄芪醇的能力[27-28]。未来,应用宏基因组学、培养组学等微生物组学方法,有望获取更多高效的微生物和糖基水解酶用于环黄芪醇的制备[29-33]。
黄芪种植占用土地,黄芪的生长易受气候、温度和湿度等多种因素影响,并且收获的黄芪中有效物质的含量不统一,难以进行规模化、标准化生产。黄芪幼苗的根中皂苷含量较高,是进行组织培养的理想部位。通过优化培养条件,可以使黄芪在组织培养过程中积累丰富的黄芪皂苷。Jiao等[34]对黄芪毛状根培养36 d后,黄芪毛状根中总生物量可达15.79 g/L(干重),总黄芪皂苷累积含量达到2.657 mg/g(干重),与田间种植3年的黄芪生根中总黄芪皂苷的含量(2.435 mg/g(干重))相近。
植物组织培养不占用土地,但需严格控制环境条件,并且培养过程中容易被污染,因此通过组织培养技术大规模获取黄芪生物质的成本较高。酸解法、Smith降解法制备环黄芪醇的成本也较高,并且需要使用多种化学试剂,生产过程易造成环境污染;酶和微生物水解法制备的环黄芪醇产物的分离纯化困难,不利于应用推广。因此,亟待开发其他绿色可持续发展的环黄芪醇制备方法。
2 环黄芪醇生物合成的关键酶黄芪植物中生物合成三萜皂苷的途径可分为以下3部分。
(1) 三萜皂苷的通用前体异戊烯基焦磷酸(isopentenyl pyrophosphate,IPP)和二甲基烯丙基焦磷酸(dimethylallyl diphosphate,DMAPP)的合成。植物具有两条合成萜类前体IPP的途径:一条是甲羟戊酸(mevalonate,MVA)途径,主要存在于植物细胞质中;另一条是2-C-甲基-D-赤藓醇-4-磷酸(2-C-methyl-D-erythritol-4-phosphate,MEP)途径,主要存在于植物细胞质体区室内。经异戊烯基焦磷酸异构酶(isopentenyl diphosphate isomerase, IDI)催化,IPP与DMAPP可实现相互转化。
(2) IPP在多种酶的催化下聚合得到2, 3-氧化角鲨烯,然后经氧化角鲨烯环化酶(2, 3-oxidosqualene cyclase,OSC)催化得到不同的三萜骨架。
(3) 三萜骨架在细胞色素P450(cytochrome P450s,CYP450s)、UDP-糖基转移酶(UDP-glycosyl transferase,UGT)等修饰下合成多种三萜类化合物。
在以上合成三萜皂苷的途径中,OSC、CYP450s等酶在合成多样化的三萜化合物方面发挥了重要作用,是植物三萜化合物生物合成中的关键酶。
2.1 氧化角鲨烯环化酶OSC是三萜皂苷下游合成阶段的第一个限速酶,可催化2, 3-氧化角鲨烯环化生成三萜骨架。Liu等[35]将雷公藤OSC基因进行了异源表达,发现TwOSC4和TwOSC6是环阿屯醇合酶。Chen等[36]从Astragalus membranaceus中克隆了两个OSC基因,并对其在膜荚黄芪中的功能进行了研究,发现AmOSC3与环阿屯醇的合成有关。Huang等[37]从Camellia sasanqua中发掘了7种OSC基因并在酵母中进行了功能表征,最终确定CsOSC5和CsOSC7为环阿屯醇合酶。Guo等[38]利用酵母表达系统确定了主要在重楼(Paris polyphylla)叶片中表达的环阿屯醇合酶基因PpCAS,这是首个报道的来自于重楼的环阿屯醇合酶基因。Kawano等[39]发现来源于Costus speciosus的CsOSC1(AB058507)基因能够编码环阿屯醇合酶,这是首个报道的单子叶植物环阿屯醇合酶。
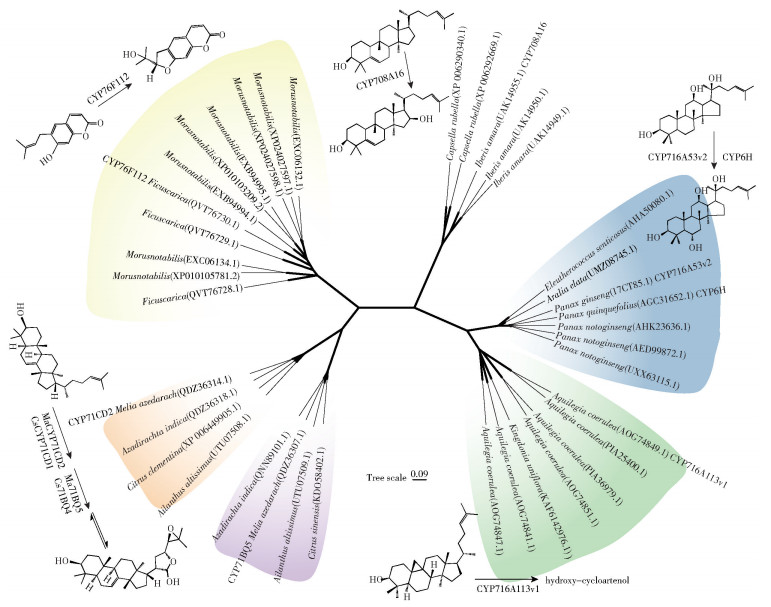
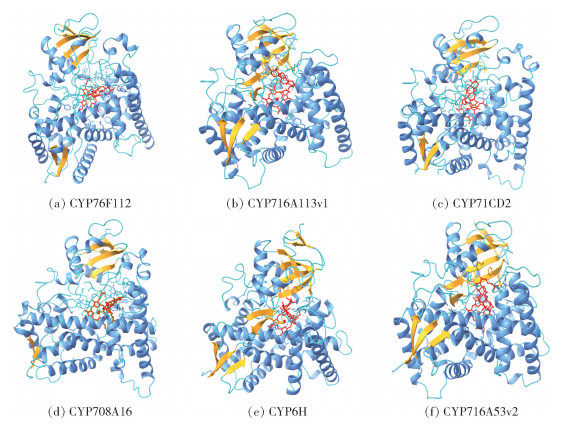
2.2 细胞色素P450单加氧酶细胞色素P450单加氧酶是一个庞大的家族,是植物次生代谢产物多样性的通用驱动因子。在三萜皂苷的合成途径中,CYP450s在底物的特定部位引入氧原子,催化三萜骨架进行羟基化、羰基化、环氧化、去甲基化等结构修饰[40-42]。通过对环阿屯醇与环黄芪醇的结构进行比较,发现环黄芪醇在C6、C16、C25位有羟基,C21与C24位环氧化为五元环,推测环阿屯醇可能经CYP450羟基化与环氧化后转变为环黄芪醇。本文分析了催化与环阿屯醇骨架相似的化合物的CYP450s,筛选了具有催化四环三萜皂苷与甾醇等的环氧化和羟基化活性的7种CYP450s,并通过比对等方法进一步从Genbank数据库中筛选了38条相关序列,以邻接法建立了系统发育树,结果见图 1。在此基础上,本文从Plant Cytochrome P450 Database、Uniprot和PubChem下载了部分CYP450s的PDB数据和环阿屯醇SDF数据,采用PCPLD和AlphaFold-P450工具(http://p450.biodesign.ac.cn/)预测了部分CYP450s与环阿屯醇的分子对接,并运用UCSF Chimerax进行了可视化处理,结果见图 2。
|
图 1 部分具有羟基化和环氧化活性的CYP450s系统发育分析 Fig.1 Phylogenetic analysis of some CYP450s with hydroxylation and epoxidation activities |
|
图 2 部分CYP450s与环阿屯醇的分子对接 Fig.2 Molecular docking between some CYP450s and cycloartenol |
人们已经对系统发育树中的几种CYP450s进行了多项酶学研究。Hodgson等[43]的研究表明,来自苦楝(Melia azedarach)的MaCYP71CD2和Ma-CYP71BQ5可氧化印楝素前体(Tirucalla-7, 24-dien-3β-ol)的尾端形成半缩醛环式结构。Villard等[44]发现CYP76F112在呋喃香豆素的合成途径中发挥着重要的环化作用。Takase等[45]对多种CYP450s在葫芦素生物合成中的作用进行了研究,他们将CYP81AQ19与葫芦二烯醇合酶在酵母中共表达,发现CYP81AQ19具有烯丙基羟基化活性;经过温和的酸处理后,可引发产物C23-OH基团异构化为C25-OH以及双键迁移。有研究表明CYP716A12和其他CYP716A亚家族酶大多为C28氧化酶[46],但部分CYP716A具有C6羟基化活性,例如CYP716A53v2可羟基化原萘二醇的C6位,得到原萘三醇[47]。将CYP6H在酿酒酵母中进行表达,也可实现外源性原萘二醇C6位的羟基化[48]。Miettinen等[49]通过酵母表征了不同药用植物的CYP716酶,他们从耧斗菜(Aquilegia coerulea)中鉴定出CYP716A113v1,该酶可能参与甾体皂苷的生物合成,能够羟化甾体皂苷前体环阿屯醇或酵母甾醇前体等其他四环三萜类化合物,但其具体氧化部位未知。Dong等[50]研究确定了葫芦素生物合成途径中的2个CYP450s,证明了CYP708A16可催化葫芦二烯醇转化为16-β-羟基葫芦二烯醇。
CYP450s在植物进化的过程中可能获得新的活性,例如苔藓植物(Calohypnum plumiforme)独立进化出2个莫内酯生物合成基因簇,它们与水稻相关酶的特性相同,但属于不同的P450家族[51]。此外,利用整合系统发育分析、蛋白质结构计算和数据驱动设计(data driven design)等方法,重新设计改造CYP87D20等多功能酶的活性位点,有望将其转变为高效的葫芦二烯醇C11羟化酶[52-53]。未来,除了从黄芪中挖掘环黄芪醇合成所需的CYP450s外,还可以通过多组学技术在多种植物中获取环黄芪醇合酶。
3 酿酒酵母底盘细胞的构建与优化酿酒酵母、大肠杆菌等微生物是异源合成多种植物天然产物的主要底盘细胞。大肠杆菌具有发酵周期短(2~3 d)、萜类合成途径简单和不分流等优势。目前,已在大肠杆菌中实现了环阿屯醇的合成[54],但是大肠杆菌缺少细胞器,CYP450s在大肠杆菌等细菌中通常难以表达[55]。此外,大肠杆菌内的MEP途径多用于合成酮类、醇类和酸类等化合物,较少用于萜类合成[56]。酵母具有MVA代谢途径,能够提供三萜合成所需的前体物质[57-58];酿酒酵母具有内质网、高尔基体、过氧化物酶体等亚细胞结构,有利于植物源关键酶基因的表达[59],并且能够为植物三萜类化合物母核的合成提供适宜的反应场所;酵母还能对萜类母核进行糖基化、羟基化等修饰。目前,已在酵母中实现了多种三萜苷元及皂苷的异源生物合成[60-63],这为酵母合成环黄芪醇提供了参考。此外,酿酒酵母也被认定为安全的微生物(generally recognized as safe, GRAS)[64],已广泛应用于多种食品、药品的合成[65-67],因此是环黄芪醇生物合成的理想宿主[68]。
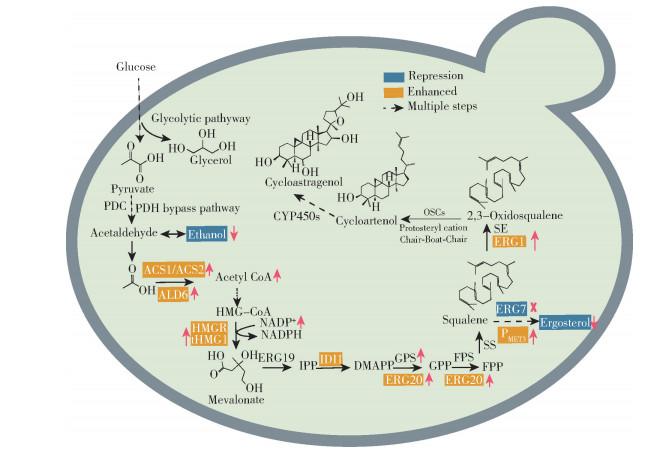
图 3显示了工程酿酒酵母中环黄芪醇的代谢途径。在酵母内源MVA途径中,葡萄糖经糖酵解途径,经乙醛脱氢酶(ALDH)、乙酰辅酶A合酶(ACS)催化后,生成重要的合成底物——乙酰辅酶A(acetyl-CoA)。乙酰辅酶A在乙酰乙酰辅酶A巯解酶(ERG10)、3-羟基-3-甲基戊二酰辅酶A合酶(ERG13)、HMG-CoA还原酶(HMGR)、甲戊二酸激酶(ERG12)、磷酸戊二酸激酶(ERG8)、法尼二磷酸合酶(ERG20)等酶的催化下反应生成IPP,经异戊烯基焦磷酸异构酶(IDI1)催化,生成DMAPP。之后,DMAPP在香叶基焦磷酸合酶(geranyl pyrophosphate synthetase,GPS)、法尼基焦磷酸合酶(farnesyl pyrophosphate synthetase,FPS)、角鲨烯合酶(squalene synthase,SS)、角鲨烯环氧酶(squalene epoxidase,SE)等的作用下,生成2, 3-氧化角鲨烯。在OSCs的作用下,2, 3-氧化角鲨烯通过“椅-船-椅”式构象变化形成环阿屯醇[69-70]。最后,环阿屯醇经CYP450s羟基化、环氧化等结构修饰,生成环黄芪醇。通过酵母生成环黄芪醇,仍需对酵母细胞进行代谢工程和合成生物学改造,以增加前体供应,并高效表达环黄芪醇合成的关键酶基因。
|
图 3 工程酿酒酵母中环黄芪醇的代谢途径 Fig.3 Metabolic pathways of cycloastragenol in engineered Sacchromyces cerevisiae |
细胞质中,乙酰辅酶A主要通过乙酰辅酶A合酶(ACS1、ACS2)途径合成[71]。在酿酒酵母中过表达ACS1和ACS2可以使乙酰辅酶A的合成产量提高2到5倍,从而增加其在细胞质中的浓度[72]。同时,乙酰辅酶A与乙醛酸生成苹果酸是细胞质和过氧化物酶体中乙醛酸循环的重要环节,抑制柠檬酸合酶(CIT2)和苹果酸合酶(MLS1)的表达可以减少乙酰辅酶A的损失[73]。酵母细胞中,同时提高乙酰和辅酶A的合成,能够大幅增加乙酰辅酶A及下游产物的产量[74]。
3.2 在细胞质中构建外源乙酰辅酶A的合成途径在酿酒酵母中引入外源乙酰辅酶A合成途径可以提高乙酰辅酶A的合成效率。Meadows等[75]使用4种非天然代谢反应重组酿酒酵母的中心碳代谢,降低了胞质乙酰辅酶A生物合成对ATP的需求和CO2的生成量,有利于经济高效地合成乙酰辅酶A。Kozak等[76]在酿酒酵母中表达乙酰乙醛脱氢酶(A-ALD),结果显示乙酰乙醛脱氢酶能够合成乙酰辅酶A。乙酰辅酶A可以在柠檬酸裂解酶(ACL)的催化下直接由柠檬酸获得,而酿酒酵母中缺少ACL基因[77]。Lian等[78]在酿酒酵母的细胞质中表达外源ACL基因,促进了乙酰辅酶A的生成。
酿酒酵母中存在运输乙酰辅酶A的(乙酰)肉毒碱穿梭系统,但是由于其无法合成肉毒碱,线粒体中的乙酰辅酶A无法被转运到细胞质中[73]。线粒体中的乙酰辅酶A合成途径消耗更少的能量[79],且乙酰辅酶A主要在线粒体中产生,因此,在细胞质中构建线粒体中的丙酮酸脱氢酶复合体具有重要意义。Lian等[80]将丙酮酸脱氢酶复合体在酵母细胞质中表达,使目标产物的产量大为提升。此外,在线粒体中构建MVA途径,具有提高乙酰辅酶A相关产物产量的潜能,例如Lv等[81]通过此方法使异戊二烯的产量达到2 527 mg/L。但是线粒体为酵母细胞能量的来源,应尽量避免在线粒体中进行萜类相关产物的合成。
3.3 解除产物对酿酒酵母的毒副作用以及改善前体合成途径以酿酒酵母为底盘细胞合成环黄芪醇,需要对酿酒酵母底盘细胞进行适当改造,消除其在合成过程中的多种限制,从而提高目标产物的产量。酿酒酵母在发酵生产过程中产生的部分代谢产物会对酿酒酵母细胞产生毒副作用,从而降低酿酒酵母的生物量。王雅楠等[82]通过在酿酒酵母中过量表达Spnox基因,使重组菌株胞内总NADH氧化酶活性提高了49%,NADH/NAD+比例降低了9%,减少了酿酒酵母中乙醇的合成,这为减少环黄芪醇合成过程中乙醇等代谢产物对酿酒酵母的影响提供了思路。
为了满足酿酒酵母细胞本身的生物代谢途径和环黄芪醇合成途径对前体物质的需要,提高关键酶基因在工程菌中的表达量是强化前体物质合成的最直接方法。HMGR是MVA途径的第一个限速酶,过表达N端截短的3-羟基-3-甲基戊二酸单酰辅酶A还原酶HMGR(tHMG1)可以促进代谢流向MVA途径转移,降低法尼基焦磷酸的反馈抑制,从而提高MVA途径中间产物的生成量[83]。在构建具有原人参二醇合成途径的酿酒酵母中,过表达编码该酶功能区的基因tHMG1后,原人参二醇产量提升为原产量的9.7倍[84];此外,过表达IDI1、ERG10、ERG20基因和角鲨烯合酶基因(ERG9)等也可以提高前体物质的供应[85]。ERG1基因的表达产物可以将角鲨烯氧化成2, 3-氧化角鲨烯,因此过表达ERG1基因可以促进2, 3-氧化角鲨烯的生成。
3.4 筛选高活性异源关键酶基因酵母细胞不具有环阿屯醇合酶、CYP450s等环黄芪醇合成所需的关键酶[86]。部分外源基因在酿酒酵母内难以表达或表达量较低[87],导致关键酶的活性不高,因此,需要筛选并优化异源基因的表达[88]。针对异源基因表达时存在的密码子偏好性问题,可以根据酿酒酵母的密码子特点,对相关基因进行密码子优化;针对异源酶活性低等问题,则可以通过定向进化、理性设计等蛋白质工程方法提高其活性和特异性[89]。酿酒酵母中缺少植物来源的酶翻译、酶修饰能力和亚细胞结构,导致植物天然产物合成所需的CYP450s等关键酶存在折叠错误、表达水平低和催化效率差等问题。未来,改善酵母底盘细胞特性,提高酵母表达CYP450s等植物天然产物合成关键酶的能力,对实现植物活性天然产物的酵母合成至关重要。
3.5 抑制羊毛甾醇竞争途径环阿屯醇合酶(CAS)可以催化2, 3-氧化角鲨烯为环黄芪醇的前体环阿屯醇,不同的氧化角鲨烯环化酶会催化2, 3-氧化角鲨烯环化成不同的化合物,并与环阿屯醇竞争前体,其中环阿屯醇合成最主要的竞争途径是羊毛甾醇的合成。酿酒酵母的ERG7基因编码的羊毛甾醇合酶可以催化2, 3-氧化角鲨烯生成羊毛甾醇,再经过一系列反应合成细胞膜中的成分麦角甾醇,从而降低环阿屯醇的产量[90-91]。由于麦角甾醇是构成细胞膜的主要成分,因此无法完全敲除ERG7基因进行调控。
启动子工程可以有效调控酵母基因的表达[92]。将ERG7基因的启动子替换为弱启动子,降低该基因的转录水平,可以减少羊毛甾醇的合成。PCTR3启动子受铜离子浓度影响,以该启动子替换酿酒酵母的ERG7基因启动子后,在培养基中添加高浓度铜离子,可显著抑制ERG7基因的转录[93]。同理,也可以利用蛋氨酸和甲硫氨酸抑制性启动子PMET3替换ERG7基因启动子,在培养基中添加蛋氨酸或甲硫氨酸,可以抑制ERG7基因的表达[94]。在酿酒酵母细胞中构建CRISPR/dCas9系统,将dCas9基因整合到酿酒酵母基因组中,表达出作用于ERG7的dCas9蛋白,也可以抑制羊毛甾醇的合成[95]。此外,还可以利用反义RNA技术抑制ERG7基因的表达[96],但是由于此技术的操作过程繁琐、成本较高,目前已很少使用。
乙酰辅酶A、法尼基焦磷酸、异戊二烯、异戊烯基焦磷酸、角鲨烯是酵母中环黄芪醇合成的重要前体或中间产物,可通过多种优化策略提高其合成产量,从而提高环阿屯醇的合成效率。目前,三萜类化合物绞股蓝皂苷、人参皂苷Rg3、人参皂苷CK、双环单萜化合物桉叶素、萜醇化合物芳樟醇、黄酮类化合物柚皮素等均在酵母中实现了高效合成(表 1),这为环黄芪醇在酵母中的高效合成提供了参考。
| 下载CSV 表 1 酿酒酵母细胞工厂高效合成策略 Table 1 Efficient synthesis strategies for cell factories in S. cerevisiae |
目前,环黄芪醇下游合成途径中CYP450s等关键酶的结构修饰的专一性较低,部分关键酶的催化机制未知,在酿酒酵母中异源表达高效关键酶基因有待进一步研究。未来,为实现环黄芪醇的酵母合成,可加强以下几方面的研究工作:
(1) 利用多组学技术和合成生物学技术,从黄芪中的环黄芪醇高效合成部位挖掘环黄芪醇合成途径的关键酶;
(2) 探究环黄芪醇在酿酒酵母中的异源合成途径,进一步筛选与酵母系统匹配的环黄芪醇合成关键酶基因;
(3) 通过启动子替换、基因敲除、基因过表达等手段,结合基因编辑技术,可以强化环黄芪醇的合成代谢途径、抑制竞争途径的代谢通量、减少中间代谢产物的积累,从而提高环黄芪醇在酵母中的合成产率、速率和得率(titer, rate and yield,TRY)。
通过合成生物学研究,有望大量获取具有市场竞争力的环黄芪醇产品,这样不仅能够实现绿色生物制造,还能够为实现碳中和、碳达峰提供帮助。
| [1] |
张蔷, 高文远, 满淑丽. 黄芪中有效成分药理活性的研究进展[J]. 中国中药杂志, 2012, 37(21): 3203-3207. ZHANG Q, GAO W Y, MAN S L. Chemical composition and pharmacological activities of Astragali Radix[J]. China Journal of Chinese Materia Medica, 2012, 37(21): 3203-3207. (in Chinese) |
| [2] |
DING W J, CHEN G H, DENG S H, et al. Calycosin protects against oxidative stress-induced cardiomyocyte apoptosis by activating aldehyde dehydrogenase 2[J]. Phytotherapy Research, 2023, 37(1): 35-49. DOI:10.1002/ptr.7591 |
| [3] |
杨冰, 于桂红, 李明雨, 等. 基于"补气固表"探究黄芪黄酮组分抑制C57BL/6荷瘤小鼠肿瘤生长及免疫调节机制研究[J]. 中国中药杂志, 2019, 44(23): 5184-5190. YANG B, YU G H, LI M Y, et al. Mechanism of flavonoid components in Astragali Radix in inhibiting tumor growth and immunoregulation in C57BL/6 tumor bearing mice based on "invigorating Qi for consolidation of exterior"[J]. China Journal of Chinese Materia Medica, 2019, 44(23): 5184-5190. (in Chinese) |
| [4] |
CHEN G H, XU H L, XU T, et al. Calycosin reduces myocardial fibrosis and improves cardiac function in post-myocardial infarction mice by suppressing TGFBR1 signaling pathways[J]. Phytomedicine, 2022, 104: 154277. DOI:10.1016/j.phymed.2022.154277 |
| [5] |
KONG X H, WANG F, NIU Y B, et al. A comparative study on the effect of promoting the osteogenic function of osteoblasts using isoflavones from Radix Astragalus[J]. Phytotherapy Research, 2018, 32(1): 115-124. DOI:10.1002/ptr.5955 |
| [6] |
LI W F, HU X Y, WANG S P, et al. Characterization and anti-tumor bioactivity of astragalus polysaccharides by immunomodulation[J]. International Journal of Biological Macromolecules, 2020, 145: 985-997. DOI:10.1016/j.ijbiomac.2019.09.189 |
| [7] |
DONG N, LI X R, XUE C Y, et al. Astragalus polysaccharides alleviates LPS-induced inflammation via the NF-κB/MAPK signaling pathway[J]. Journal of Cellular Physiology, 2020, 235(7-8): 5525-5540. DOI:10.1002/jcp.29452 |
| [8] |
LIU T L, ZHANG M J, NIU H Y, et al. Astragalus polysaccharide from Astragalus Melittin ameliorates inflammation via suppressing the activation of TLR-4/NF-κB p65 signal pathway and protects mice from CVB3-induced virus myocarditis[J]. International Journal of Biological Macromolecules, 2019, 126: 179-186. DOI:10.1016/j.ijbiomac.2018.12.207 |
| [9] |
SU H F, SHAKER S, KUANG Y, et al. Phytochemistry and cardiovascular protective effects of Huang-Qi (Astragali Radix)[J]. Medicinal Research Reviews, 2021, 41(4): 1999-2038. DOI:10.1002/med.21785 |
| [10] |
LI M, LI S C, DOU B K, et al. Cycloastragenol upregulates SIRT1 expression, attenuates apoptosis and suppresses neuroinflammation after brain ischemia[J]. Acta Pharmacologica Sinica, 2020, 41(8): 1025-1032. DOI:10.1038/s41401-020-0386-6 |
| [11] |
IKRAM M, JO M H, CHOE K, et al. Cycloastragenol, a triterpenoid saponin, regulates oxidative stress, neurotrophic dysfunctions, neuroinflammation and apoptotic cell death in neurodegenerative conditions[J]. Cells, 2021, 10(10): 2719. DOI:10.3390/cells10102719 |
| [12] |
WAN Y, XU L, WANG Y X, et al. Preventive effects of astragaloside Ⅳ and its active sapogenin cycloastragenol on cardiac fibrosis of mice by inhibiting the NLRP3 inflammasome[J]. European Journal of Pharmacology, 2018, 833: 545-554. DOI:10.1016/j.ejphar.2018.06.016 |
| [13] |
DENG G L, ZHOU L S, WANG B L, et al. Targeting cathepsin B by cycloastragenol enhances antitumor immunity of CD8 T cells via inhibiting MHC-I degradation[J]. Journal for ImmunoTherapy of Cancer, 2022, 10(10): e004874. DOI:10.1136/jitc-2022-004874 |
| [14] |
张阳焕, 袁洋, 孙曼婷, 等. 环黄芪醇抗衰老药理作用的研究进展[J]. 中国细胞生物学学报, 2021, 43(10): 2078-2084. ZHANG Y H, YUAN Y, SUN M T, et al. Research progress of anti-aging pharmacological effect of cycloastragalol[J]. Chinese Journal of Cell Biology, 2021, 43(10): 2078-2084. (in Chinese) |
| [15] |
CRAVENS A, PAYNE J, SMOLKE C D. Synthetic biology strategies for microbial biosynthesis of plant natural products[J]. Nature Communications, 2019, 10(1): 2142. DOI:10.1038/s41467-019-09848-w |
| [16] |
ZHU M, WANG C X, SUN W T, et al. Boosting 11-oxo-β-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants[J]. Metabolic Engineering, 2018, 45: 43-50. DOI:10.1016/j.ymben.2017.11.009 |
| [17] |
YAN X, FAN Y, WEI W, et al. Production of bioactive ginsenoside compound K in metabolically engineered yeast[J]. Cell Research, 2014, 24(6): 770-773. DOI:10.1038/cr.2014.28 |
| [18] |
WANG P P, WEI Y J, FAN Y, et al. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts[J]. Metabolic Engineering, 2015, 29: 97-105. DOI:10.1016/j.ymben.2015.03.003 |
| [19] |
WEI W, WANG P P, WEI Y J, et al. Characterization of Panax ginseng UDP-glycosyltransferases catalyzing protopanaxatriol and biosyntheses of bioactive ginsenosides F1 and Rh1 in metabolically engineered yeasts[J]. Molecular Plant, 2015, 8(9): 1412-1424. DOI:10.1016/j.molp.2015.05.010 |
| [20] |
MOSES T, POLLIER J, ALMAGRO L, et al. Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum[J]. Proceedings of the National Academy of Sciences, 2014, 111(4): 1634-1639. DOI:10.1073/pnas.1323369111 |
| [21] |
ITKIN M, DAVIDOVICH-RIKANATI R, COHEN S, et al. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside Ⅴ from Siraitia grosvenorii[J]. Proceedings of the National Academy of Sciences, 2016, 113(47): E7619-E7628. |
| [22] |
楚治良, 王好锋, 韩静, 等. 中药单体环黄芪醇的制备方法[J]. 实用医药杂志, 2019, 36(9): 822-824. CHU Z L, WANG H F, HAN J, et al. Study on the preparation of TCM cycloastragenol[J]. Practical Journal of Medicine & Pharmacy, 2019, 36(9): 822-824. (in Chinese) |
| [23] |
FENG L M, LIN X H, HUANG F X, et al. Smith degradation, an efficient method for the preparation of cycloastragenol from astragaloside Ⅳ[J]. Fitoterapia, 2014, 95: 42-50. DOI:10.1016/j.fitote.2014.02.014 |
| [24] |
CHENG L Y, ZHANG H, LIANG H, et al. Enzymatic bioconversion of cycloastragenol-6-O-β-D-glucoside into cycloastragenol by a novel recombinant β-glucosidase from Phycicoccus sp. Soil748[J]. Process Biochemistry, 2020, 90: 81-88. DOI:10.1016/j.procbio.2019.11.006 |
| [25] |
LI Q, WU T, ZHAO L G, et al. Highly efficient biotransformation of astragaloside Ⅳ to cycloastragenol by sugar-stimulated β-glucosidase and β-xylosidase from Dictyoglomus thermophilum[J]. Journal of Microbiology and Biotechnology, 2019, 29(12): 1882-1893. DOI:10.4014/jmb.1807.07020 |
| [26] |
CHENG L Y, ZHANG H, CUI H Y, et al. Efficient production of the anti-aging drug cycloastragenol: insight from two glycosidases by enzyme mining[J]. Applied Microbiology and Biotechnology, 2020, 104(23): 9991-10004. DOI:10.1007/s00253-020-10966-5 |
| [27] |
WANG L M, CHEN Y. Efficient biotransformation of astragaloside Ⅳ to cycloastragenol by Bacillus sp. LG-502[J]. Applied Biochemistry and Biotechnology, 2017, 183(4): 1488-1502. DOI:10.1007/s12010-017-2517-1 |
| [28] |
TAKEUCHI D M, KISHINO S, OZEKI Y, et al. Analysis of astragaloside Ⅳ metabolism to cycloastragenol in human gut microorganism, bifidobacteria, and lactic acid bacteria[J]. Bioscience, Biotechnology, and Biochemistry, 2022, 86(10): 1467-1475. DOI:10.1093/bbb/zbac130 |
| [29] |
LIANG J W, MAI W N, WANG J, et al. Performance and microbial communities of a novel integrated industrial-scale pulp and paper wastewater treatment plant[J]. Journal of Cleaner Production, 2021, 278: 123896. DOI:10.1016/j.jclepro.2020.123896 |
| [30] |
WANG J, LIANG J W, LI Y H, et al. Characterization of efficient xylanases from industrial-scale pulp and paper wastewater treatment microbiota[J]. AMB Express, 2021, 11: 19. DOI:10.1186/s13568-020-01178-1 |
| [31] |
魏勇军, 李晓琪, 戢博阳, 等. 肠道菌群与宿主关系解析及肠道菌群调控/合成研究进展[J]. 中国科学: 生命科学, 2022, 52(2): 249-265. WEI Y J, LI X Q, JI B Y, et al. Recent advances on the recovery, modulation and synthetic biology of gut microbiota and hosts[J]. Scientia Sinica(Vitae), 2022, 52(2): 249-265. (in Chinese) |
| [32] |
MIAO Q, ZHANG X L, WANG Y T, et al. Characterization of novel pectinolytic enzymes derived from the efficient lignocellulose degradation microbiota[J]. Biomolecules, 2022, 12(10): 1388. DOI:10.3390/biom12101388 |
| [33] |
YANG Y D, QU L B, MIJAKOVIC I, et al. Advances in the human skin microbiota and its roles in cutaneous diseases[J]. Microbial Cell Factories, 2022, 21(1): 176. DOI:10.1186/s12934-022-01901-6 |
| [34] |
JIAO J, GAI Q Y, FU Y J, et al. Optimization of Astragalus membranaceus hairy roots induction and culture conditions for augmentation production of astragalosides[J]. Plant Cell, Tissue and Organ Culture, 2015, 120: 1117-1130. DOI:10.1007/s11240-014-0668-0 |
| [35] |
LIU Y, ZHOU J W, HU T Y, et al. Identification and functional characterization of squalene epoxidases and oxidosqualene cyclases from Tripterygium wilfordii[J]. Plant Cell Reports, 2020, 39(3): 409-418. DOI:10.1007/s00299-019-02499-7 |
| [36] |
CHEN K, ZHANG M, XU L L, et al. Identification of oxidosqualene cyclases associated with saponin biosynthesis from Astragalus membranaceus reveals a conserved motif important for catalytic function[J]. Journal of Advanced Research, 2023, 43: 247-257. DOI:10.1016/j.jare.2022.03.014 |
| [37] |
HUANG L F, HU Y E, HUANG R S, et al. Oxidosqualene cyclases involved in the biosynthesis of diverse triterpenes in Camellia sasanqua[J]. Journal of Agricultural and Food Chemistry, 2022, 70(26): 8075-8084. DOI:10.1021/acs.jafc.2c03011 |
| [38] |
GUO S Y, YIN Y, LEI T, et al. A cycloartenol synthase from the steroidal saponin biosynthesis pathway of Paris polyphylla[J]. Journal of Asian Natural Products Research, 2021, 23(4): 353-362. DOI:10.1080/10286020.2020.1730331 |
| [39] |
KAWANO N, ICHINOSE K, EBIZUKA Y. Molecular cloning and functional expression of cDNAs encoding oxidosqualene cyclases from Costus speciosus[J]. Biological and Pharmaceutical Bulletin, 2002, 25(4): 477-482. DOI:10.1248/bpb.25.477 |
| [40] |
GIRVAN H M, MUNRO A W. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology[J]. Current Opinion in Chemical Biology, 2016, 31: 136-145. DOI:10.1016/j.cbpa.2016.02.018 |
| [41] |
GUENGERICH F P, MUNRO A W. Unusual cytochrome P450 enzymes and reactions[J]. Journal of Biological Chemistry, 2013, 288(24): 17065-17073. DOI:10.1074/jbc.R113.462275 |
| [42] |
MALHOTRA K, FRANKE J. Cytochrome P450 monooxygenase-mediated tailoring of triterpenoids and steroids in plants[J]. Beilstein Journal of Organic Chemistry, 2022, 18: 1289-1310. DOI:10.3762/bjoc.18.135 |
| [43] |
HODGSON H, DE LA PEÑA R, STEPHENSON M J, et al. Identification of key enzymes responsible for protolimonoid biosynthesis in plants: opening the door to azadirachtin production[J]. Proceedings of the National Academy of Sciences, 2019, 116(34): 17096-17104. DOI:10.1073/pnas.1906083116 |
| [44] |
VILLARD C, MUNAKATA R, KITAJIMA S, et al. A new P450 involved in the furanocoumarin pathway underlies a recent case of convergent evolution[J]. New Phytologist, 2021, 231(5): 1923-1939. DOI:10.1111/nph.17458 |
| [45] |
TAKASE S, KERA K, NAGASHIMA Y, et al. Allylic hydroxylation of triterpenoids by a plant cytochrome P450 triggers key chemical transformations that produce a variety of bitter compounds[J]. Journal of Biological Chemistry, 2019, 294(49): 18662-18673. DOI:10.1074/jbc.RA119.009944 |
| [46] |
YASUMOTO S, FUKUSHIMA E O, SEKI H, et al. Novel triterpene oxidizing activity of Arabidopsis thaliana CYP716A subfamily enzymes[J]. FEBS Letters, 2016, 590(4): 533-540. DOI:10.1002/1873-3468.12074 |
| [47] |
HAN J Y, HWANG H S, CHOI S W, et al. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng[J]. Plant and Cell Physiology, 2012, 53(9): 1535-1545. DOI:10.1093/pcp/pcs106 |
| [48] |
WANG L, ZHAO S J, LIANG Y L, et al. Identification of the protopanaxatriol synthase gene CYP6H for ginsenoside biosynthesis in Panax quinquefolius[J]. Functional & Integrative Genomics, 2014, 14(3): 559-570. |
| [49] |
MIETTINEN K, POLLIER J, BUYST D, et al. The ancient CYP716 family is a major contributor to the diversification of eudicot triterpenoid biosynthesis[J]. Nature Communications, 2017, 8: 14153. DOI:10.1038/ncomms14153 |
| [50] |
DONG L, ALMEIDA A, POLLIER J, et al. An independent evolutionary origin for insect deterrent cucurbitacins in Iberis amara[J]. Molecular Biology and Evolution, 2021, 38(11): 4659-4673. DOI:10.1093/molbev/msab213 |
| [51] |
ZHANG J, PETERS R J. Why are momilactones always associated with biosynthetic gene clusters in plants?[J]. Proceedings of the National Academy of Sciences, 2020, 117(25): 13867-13869. DOI:10.1073/pnas.2007934117 |
| [52] |
LI D W, MA Y S, ZHOU Y, et al. A structural and data-driven approach to engineering a plant cytochrome P450 enzyme[J]. Science China Life Sciences, 2019, 62(7): 873-882. DOI:10.1007/s11427-019-9538-3 |
| [53] |
ZHANG X P, LUO W, YAO Y Y, et al. Enhanced chemoselectivity of a plant cytochrome P450 through protein engineering of surface and catalytic residues[J]. aBIOTECH, 2021, 2(3): 215-225. DOI:10.1007/s42994-021-00056-z |
| [54] |
TAKEMURA M, TANAKA R, MISAWA N. Pathway engineering for the production of β-amyrin and cycloartenol in Escherichia coli—a method to biosynthesize plant-derived triterpene skeletons in E. coli[J]. Applied Microbiology and Biotechnology, 2017, 101(17): 6615-6625. DOI:10.1007/s00253-017-8409-z |
| [55] |
AJIKUMAR P K, XIAO W H, TYO K E J, et al. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli[J]. Science, 2010, 330(6000): 70-74. DOI:10.1126/science.1191652 |
| [56] |
JARBOE L R, ZHANG X, WANG X, et al. Metabolic engineering for production of biorenewable fuels and chemicals: contributions of synthetic biology[J]. Journal of Biomedicine and Biotechnology, 2010, 2010: 761042. |
| [57] |
KAMPRANIS S C, MAKRIS A M. Developing a yeast cell factory for the production of terpenoids[J]. Computational and Structural Biotechnology Journal, 2012, 3(4): e201210006. DOI:10.5936/csbj.201210006 |
| [58] |
李冰, 张传波, 宋凯, 等. 生物合成稀有人参皂苷的研究进展[J]. 中国生物工程杂志, 2021, 41(6): 71-88. LI B, ZHANG C B, SONG K, et al. Research progress in biosynthesis of rare ginsenosides[J]. China Biotechnology, 2021, 41(6): 71-88. (in Chinese) |
| [59] |
陈明凯, 叶丽丹, 于洪巍. 代谢改造酿酒酵母合成萜类化合物的研究进展[J]. 生物工程学报, 2021, 37(6): 2085-2104. CHEN M K, YE L D, YU H W. Advances in metabolic engineering of Saccharomyces cerevisiae for terpenoids biosynthesis[J]. Chinese Journal of Biotechnology, 2021, 37(6): 2085-2104. (in Chinese) |
| [60] |
YANG C S, LI C J, WEI W, et al. The unprecedented diversity of UGT94-family UDP-glycosyltransferases in Panax plants and their contribution to ginsenoside biosynthesis[J]. Scientific Reports, 2020, 10(1): 15394. DOI:10.1038/s41598-020-72278-y |
| [61] |
PENG J J, WANG L, WANG M G, et al. Yeast synthetic biology for the production of Lycium barbarum polysaccharides[J]. Molecules, 2021, 26(6): 1641. DOI:10.3390/molecules26061641 |
| [62] |
WEI Y J, JI B Y, LEDESMA-AMARO R, et al. Editorial: engineering yeast to produce plant natural products[J]. Frontiers in Bioengineering and Biotechnology, 2021, 9: 798097. DOI:10.3389/fbioe.2021.798097 |
| [63] |
WEI Y J. Yeast synthetic biology for the production of terpenoids derived from traditional Chinese medicinal plants[M]//HARZEVILI F D. Synthetic biology of yeasts: tools and applications. Cham: Springer International Publishing, 2022: 181-205.
|
| [64] |
PARAPOULI M, VASILEIADIS A, AFENDRA A S, et al. Saccharomyces cerevisiae and its industrial applications[J]. AIMS Microbiology, 2020, 6(1): 1-31. DOI:10.3934/microbiol.2020001 |
| [65] |
WEI Y J, GOSSING M, BERGENHOLM D, et al. Increasing cocoa butter-like lipid production of Saccharomyces cerevisiae by expression of selected cocoa genes[J]. AMB Express, 2017, 7(1): 34. DOI:10.1186/s13568-017-0333-1 |
| [66] |
WEI Y J, SIEWERS V, NIELSEN J. Cocoa butter-like lipid production ability of non-oleaginous and oleaginous yeasts under nitrogen-limited culture conditions[J]. Applied Microbiology and Biotechnology, 2017, 101(9): 3577-3585. DOI:10.1007/s00253-017-8126-7 |
| [67] |
BERGENHOLM D, GOSSING M, WEI Y J, et al. Modulation of saturation and chain length of fatty acids in Saccharomyces cerevisiae for production of cocoa butter-like lipids[J]. Biotechnology and Bioengineering, 2018, 115(4): 932-942. DOI:10.1002/bit.26518 |
| [68] |
石玉松, 王冬, 李荣生, 等. 创建酿酒酵母细胞工厂发酵生产人参皂苷Rh2[J]. 中国中药杂志, 2022, 47(3): 651-658. SHI Y S, WANG D, LI R S, et al. Construction of cell factories for high production of ginsenoside Rh2 in Saccharomyces cerevisiae[J]. China Journal of Chinese Materia Medica, 2022, 47(3): 651-658. (in Chinese) |
| [69] |
陈翠玉, 庞亚如, 陈泉冰, 等. 环氧角鲨烯环化酶在三萜化合物生物合成中的进展[J]. 生物工程学报, 2022, 38(2): 443-459. CHEN C Y, PANG Y R, CHEN Q B, et al. Oxidosqualene cyclases in triterpenoids biosynthesis: a review[J]. Chinese Journal of Biotechnology, 2022, 38(2): 443-459. (in Chinese) |
| [70] |
徐圆圆, 陈仲, 贾黎明, 等. 植物三萜皂苷生物合成途径及调控机制研究进展[J]. 中国科学: 生命科学, 2021, 51(5): 525-555. XU Y Y, CHEN Z, JIA L M, et al. Advances in understanding of the biosynthetic pathway and regulatory mechanism of triterpenoid saponins in plants[J]. Scientia Sinica(Vitae), 2021, 51(5): 525-555. (in Chinese) |
| [71] |
SHIBA Y, PARADISE E M, KIRBY J, et al. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids[J]. Metabolic Engineering, 2007, 9: 160-168. DOI:10.1016/j.ymben.2006.10.005 |
| [72] |
陈孚江, 周景文, 史仲平, 等. 乙酰辅酶A合成代谢对酿酒酵母生理功能的影响[J]. 微生物学报, 2010, 50(9): 1172-1179. CHEN F J, ZHOU J W, SHI Z P, et al. Effect of acetyl-CoA synthase gene overexpression on physiological function of Saccharomyces cerevisiae[J]. Acta Microbiologica Sinica, 2010, 50(9): 1172-1179. (in Chinese) |
| [73] |
CHEN Y, DAVIET L, SCHALK M, et al. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism[J]. Metabolic Engineering, 2013, 15: 48-54. DOI:10.1016/j.ymben.2012.11.002 |
| [74] |
LIU W, ZHANG B, JIANG R. Improving acetyl-CoA biosynthesis in Saccharomyces cerevisiae via the overexpression of pantothenate kinase and PDH bypass[J]. Biotechnology for Biofuels, 2017, 10: 41. DOI:10.1186/s13068-017-0726-z |
| [75] |
MEADOWS A L, HAWKINS K M, TSEGAYE Y, et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production[J]. Nature, 2016, 537(7622): 694-697. DOI:10.1038/nature19769 |
| [76] |
KOZAK B U, VAN ROSSUM H M, BENJAMIN K R, et al. Replacement of the Saccharomyces cerevisiae acetyl-CoA synthetases by alternative pathways for cytosolic acetyl-CoA synthesis[J]. Metabolic Engineering, 2014, 21: 46-59. DOI:10.1016/j.ymben.2013.11.005 |
| [77] |
HYNES M J, MURRAY S L. ATP-citrate lyase is required for production of cytosolic acetyl coenzyme A and development in Aspergillus nidulans[J]. Eukaryotic Cell, 2010, 9(7): 1039-1048. DOI:10.1128/EC.00080-10 |
| [78] |
LIAN J, SI T, NAIR N U, et al. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains[J]. Metabolic Engineering, 2014, 24: 139-149. DOI:10.1016/j.ymben.2014.05.010 |
| [79] |
VICKERS C E, WILLIAMS T C, PENG B, et al. Recent advances in synthetic biology for engineering isoprenoid production in yeast[J]. Current Opinion in Chemical Biology, 2017, 40: 47-56. DOI:10.1016/j.cbpa.2017.05.017 |
| [80] |
LIAN J, ZHAO H. Functional reconstitution of a pyruvate dehydrogenase in the cytosol of Saccharomyces cerevisiae through lipoylation machinery engineering[J]. ACS Synthetic Biology, 2016, 5(7): 689-697. DOI:10.1021/acssynbio.6b00019 |
| [81] |
LV X M, WANG F, ZHOU P P, et al. Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae[J]. Nature Communications, 2016, 7(1): 12851. DOI:10.1038/ncomms12851 |
| [82] |
王雅楠, 宋育阳, 刘延琳, 等. 过量表达NADH氧化酶降低酿酒酵母乙醇合成的研究[J]. 中国食品学报, 2018, 18(4): 23-29. WANG Y N, SONG Y Y, LIU Y L, et al. Reduction of ethanol yield by NADH oxidase overexpression in Saccharomyces cerevisiae[J]. Journal of Chinese Institute of Food Science and Technology, 2018, 18(4): 23-29. (in Chinese) |
| [83] |
高扬乐, 谢梦斯, 李力. 利用不同底盘细胞开展生物合成萜类化合物的研究进展[J]. 药物生物技术, 2022, 29(1): 95-101. GAO Y L, XIE M S, LI L. Research progress in biosynthesis of terpenoids using different chassis cells[J]. Pharmaceutical Biotechnology, 2022, 29(1): 95-101. (in Chinese) |
| [84] |
梁会超, 胡宗风, 梁兰, 等. 过表达HMGR催化结构域以优化酵母工程菌原人参二醇代谢途径的研究[J]. 药学研究, 2016, 35(8): 444-448, 493. LIANG H C, HU Z F, LIANG L, et al. Optimization of the protopanoxadiol metabolic pathway by overexpression of the catalytic domain of HMGR in Saccharomyces cerevisiae[J]. Journal of Pharmaceutical Research, 2016, 35(8): 444-448, 493. (in Chinese) |
| [85] |
WESTFALL P J, PITERA D J, LENIHAN J R, et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin[J]. Proceedings of the National Academy of Sciences, 2012, 109(3): E111-E111. |
| [86] |
SUN J C, XU X, XUE Z Y, et al. Functional analysis of a rice oxidosqualene cyclase through total gene synthesis[J]. Molecular Plant, 2013, 6(5): 1726-1729. DOI:10.1093/mp/sst038 |
| [87] |
于欣水, 曾钰, 李洁, 等. MHF组蛋白折叠复合体组分编码基因MHF1过表达对重组酿酒酵母纤维素酶生产的影响[J]. 微生物学通报, 2019, 46(1): 75-83. YU X S, ZENG Y, LI J, et al. Effects of overexpression of the MHF histone fold complex component encoding gene MHF1 on production of cellulase by the recombinant Saccharomyces cerevisiae[J]. Microbiology China, 2019, 46(1): 75-83. (in Chinese) |
| [88] |
WANG M G, WEI Y J, JI B Y, et al. Advances in metabolic engineering of Saccharomyces cerevisiae for cocoa butter equivalent production[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 594081. |
| [89] |
GUAN R B, WANG M G, GUAN Z H, et al. Metabolic engineering for glycyrrhetinic acid production in Saccharomyces cerevisiae[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 588255. |
| [90] |
朱明. 酿酒酵母合成甘草次酸的途径构建与调控[D]. 天津: 天津大学, 2017. ZHU M. Construction and regulation of glycyrrhetinic acid synthetic pathway in Saccharomyces cerevisiae[D]. Tianjin: Tianjin University, 2017. (in Chinese) |
| [91] |
曹龙辉, 李晓珺, 赵文红, 等. 麦角甾醇的研究进展[J]. 中国酿造, 2014, 33(4): 9-12. CAO L H, LI X J, ZHAO W H, et al. Research progress on ergosterol[J]. China Brewing, 2014, 33(4): 9-12. (in Chinese) |
| [92] |
TANG H T, WU Y L, DENG J L, et al. Promoter architecture and promoter engineering in Saccharomyces cerevisiae[J]. Metabolites, 2020, 10(8): 320. |
| [93] |
ASADOLLAHI M A, MAURY J, MØLLER K, et al. Production of plant sesquiterpenes in Saccharomyces cerevisiae: effect of ERG9 repression on sesquiterpene biosynthesis[J]. Biotechnology and Bioengineering, 2008, 99(3): 666-677. |
| [94] |
KIRBY J, ROMANINI D W, PARADISE E M, et al. Engineering triterpene production in Saccharomyces cerevisiae-β-amyrin synthase from Artemisia annua[J]. The FEBS Journal, 2008, 275(8): 1852-1859. |
| [95] |
JENSEN E D, FERREIRA R, JAKOČŪNAS T, et al. Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies[J]. Microbial Cell Factories, 2017, 16(1): 46. |
| [96] |
王庆华, 高丽丽, 梁会超, 等. 利用反义RNA技术抑制酿酒酵母羊毛甾醇合酶基因的表达[J]. 药学学报, 2015, 50(1): 118-122. WANG Q H, GAO L L, LIANG H C, et al. Downregulation of lanosterol synthase gene expression by antisense RNA technology in Saccharomyces cerevisiae[J]. Acta Pharmaceutica Sinica, 2015, 50(1): 118-122. (in Chinese) |
| [97] |
郭俊琪, 王征, 张伟欣, 等. 代谢工程改造酿酒酵母提高法尼醇产量[J]. 微生物学报, 2021, 61(5): 1257-1268. GUO J Q, WANG Z, ZHANG W X, et al. Metabolic engineering of Saccharomyces cerevisiae to improve farnesol production[J]. Acta Microbiologica Sinica, 2021, 61(5): 1257-1268. (in Chinese) |
| [98] |
DAI Z B, WANG B B, LIU Y, et al. Producing aglycons of ginsenosides in bakers' yeast[J]. Scientific Reports, 2014, 4: 3698. |
| [99] |
JIANG Z Q, GAO H Y, LIU R, et al. Key glycosyltransferase genes of Panax notoginseng: identification and engineering yeast construction of rare ginsenosides[J]. ACS Synthetic Biology, 2022, 11(7): 2394-2404. |
| [100] |
SHI Y S, WANG D, LI R S, et al. Engineering yeast subcellular compartments for increased production of the lipophilic natural products ginsenosides[J]. Metabolic Engineering, 2021, 67: 104-111. |
| [101] |
IGNEA C, CVETKOVIC I, LOUPASSAKI S, et al. Improving yeast strains using recyclable integration cassettes, for the production of plant terpenoids[J]. Microbial Cell Factories, 2011, 10: 4. |
| [102] |
孙明雪, 刘继栋, 堵国成, 等. 调控酿酒酵母类异戊二烯合成途径强化芳樟醇合成[J]. 生物工程学报, 2013, 29(6): 751-759. SUN M X, LIU J D, DU G C, et al. Regulation of isoprenoid pathway for enhanced production of linalool in Saccharomyces cerevisiae[J]. Chinese Journal of Biotechnology, 2013, 29(6): 751-759. (in Chinese) |








